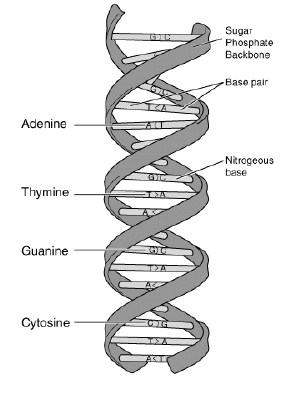
Figure 1.
DNA (Deoxyribonucleic Acid) is the molecule which contains the genetic instructions. It consists of four different nucleotides, namely Adenine, Thymine, Guanine, and Cytosine as shown in Figure 1.
If we represent a nucleotide by its initial character, a DNA strand can be regarded as a long string (sequence of characters) consisting of the four characters A, T, G, and C. For example, assume we are given some part of a DNA strand which is composed of
the following sequence of nucleotides:
``Thymine-Adenine-Adenine-Cytosine-Thymine-Guanine-Cytosine-Cytosine-Guanine-Adenine-Thymine"
Then we can represent the above DNA strand with the string ``TAACTGCCGAT." The biologist Prof. Ahn found that a gene X commonly exists in the DNA strands of five different kinds of animals, namely dogs, cats, horses, cows, and monkeys. He also discovered that
the DNA sequences of the gene X from each animal were very alike. See Figure 2.
|
DNA sequence of gene X |
| Cat: |
GCATATGGCTGTGCA |
| Dog: |
GCAAATGGCTGTGCA |
| Horse: |
GCTAATGGGTGTCCA |
| Cow: |
GCAAATGGCTGTGCA |
| Monkey: |
GCAAATCGGTGAGCA |
Figure 2. DNA sequences of gene X in five animals.
Prof. Ahn thought that humans might also have the gene X and decided to search for the DNA sequence of X in human DNA. However, before searching, he should define a representative DNA sequence of gene X because its sequences are not exactly the same in the
DNA of the five animals. He decided to use the Hamming distance to define the representative sequence. The Hamming distance is the number of different characters at each position from two strings of equal length. For example, assume we are given the two strings
``AGCAT" and ``GGAAT." The Hamming distance of these two strings is 2 because the 1st and the 3rd characters of the two strings are different. Using the Hamming distance, we can define a representative string for a set of multiple strings
of equal length. Given a set of stringsS=s1,...,smof lengthn, the consensus error between a stringyof
lengthnand the setSis the sum of the Hamming distances betweenyand eachsiinS.
If the consensus error betweenyandSis the minimum among all possible stringsyof lengthn,yis
called a consensus string ofS. For example, given the three strings ``AGCAT" ``AGACT" and ``GGAAT" the consensus string of the given strings is ``AGAAT" because the sum of the Hamming
distances between ``AGAAT" and the three strings is 3 which is minimal. (In this case, the consensus string is unique, but in general, there can be more than one consensus string.) We use the consensus string as a representative of the DNA sequence.
For the example of Figure 2 above, a consensus string of gene X is ``GCAAATGGCTGTGCA" and the consensus error is 7.
Your program is to read from standard input. The input consists ofTtest
cases. The number of test cases
Tis given in the first line of the input. Each test case starts with a line containing two integers
mand
nwhich are
separated by a single space. The integer
m(4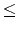 m
m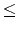 50)
50)represents
the number of DNA sequences and
n(4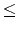 n
n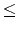 1000)
1000)represents
the length of the DNA sequences, respectively. In each of the next
mlines, each DNA sequence is given.
Your program is to write to standard output. Print the consensus string in the first line of each case and the consensus error in the second line of each case. If there exists more than one consensus string, print the lexicographically smallest consensus string.
The following shows sample input and output for three test cases.
3
5 8
TATGATAC
TAAGCTAC
AAAGATCC
TGAGATAC
TAAGATGT
4 10
ACGTACGTAC
CCGTACGTAG
GCGTACGTAT
TCGTACGTAA
6 10
ATGTTACCAT
AAGTTACGAT
AACAAAGCAA
AAGTTACCTT
AAGTTACCAA
TACTTACCAA
TAAGATAC
7
ACGTACGTAA
6
AAGTTACCAA
12
题意:给定n个DNA,求一个目标串,使得每个序列变换成目标所需次数最少。
思路:贪心。每一位选存在最多的变化即可。
代码:
#include <stdio.h>
#include <string.h>
const int N = 1005;
const int M = 55;
int t, n, m, vis[500], ans;
char g[M][N];
void init() {
ans = 0;
scanf("%d%d", &n, &m);
for (int i = 0; i < n; i ++)
scanf("%s", g[i]);
}
char cal(int i) {
char c; int Max = 0; memset(vis, 0, sizeof(vis));
for (int j = 0; j < n; j ++) {
vis[g[j][i]] ++;
if (Max < vis[g[j][i]]) {
Max = vis[g[j][i]];
c = g[j][i];
}
else if (Max == vis[g[j][i]] && g[j][i] < c) {
c = g[j][i];
}
}
ans += (n - vis[c]);
return c;
}
void solve() {
for (int i = 0; i < m; i ++)
printf("%c", cal(i));
printf("\n%d\n", ans);
}
int main() {
scanf("%d", &t);
while (t--) {
init();
solve();
}
return 0;
}
分享到:




相关推荐
连续多智能体系统的有限时间一致性问题,苗国英,徐胜元,本文分别研究了一阶和二阶多智能体系统有限时间一致性问题,其中利用了相关位置和速度的信息。当领导者的速度不能测量时,对于一阶�
Decentralized event-triggered consensus control for second-order multi-agent systems
藏经阁-Bnng Consensus to Data Replica.pdf
藏经阁-Bring Consensus to Date Replication.pdf
Fast finite-time consensus tracking of second-order multi-agent systems with a virtual leader
藏经阁-Bnng Consensus to Data Replica (1).pdf
属性图的自监督一致性表示学习_Self-supervised Consensus Representation Learning for Attributed Graph.pdf
Leader-following consensus problem with an accelerated motion leader
两流共识网络提交HACS挑战2021弱监督学习轨迹_Two-Stream Consensus Network Submission to HACS Challenge 2021 Weakly-Supervised Learning Track.pdf
DB - Just Say NO to Paxos Overhead- Replacing Consensus with Network Ordering.pdf Distributed applications use replication, implemented by protocols like Paxos, to ensure data availability and ...
Finite-time consensus for multi-agent systems via nonlinear control protocols
Finite-time consensus of multiagent systems with a switching protocol
Leader-following consensus of nonlinear multiagent Systems With Stochastic Sampling
Fully distributed finite-time consensus of directed multiquadcopter systems via pinning control
Discontinuous observers design for finite-time consensus of multiagent systems with external disturbances
In this paper a leader-following consensus problem of second-order multi-agent systems with fixed and switching topologies as well as non-uniform time-varying delays is considered For the case of ...
事件触发通信下的多智能体系统均值一致,樊渊,,本文研究了多智能体系统在事件触发控制和通信下的均值一致问题。事件触发一致性控制中,每个智能体的事件仅当状态测量的误差超出��
有向通信拓扑下多自主体系统非线性一致性协议设计,许耀赆,田玉平,本文研究了有向通信拓扑下多自主体系统的一致性问题,提出了一种设计和分析非线性一致性协议的框架。首先,通过有向图分解揭示了信�
具有通信时延的离散时间二阶多自主体系统的补偿一致性算法,刘成林,,针对具有通信时延的离散时间二阶多自主体系统系统的动态一致性问题,本文通过在常规异步耦合一致性算法中引入邻居状态状态补偿来��
在不确定扰动情况下多智能体系统的自适应---脉冲一致性控制研究,郭万里,,本文研究了多智能体系统在不确定扰动情况下的一致性问题。为使系统达到最终的一致,文章中设计了一种自适应---脉冲控制协议。...